Cell image analysis
This project is carried out in collaboration with the Faculty of Biology of Moscow State University. We analyse images of cell cultures, detect budding cells and parse structure of cellular tissue. Automatic analysis of cell images and monitoring their state alleviates work on biological experiments greatly. It saves lots of time and effort for researchers.
Monitoring cell division intensity in budding yeast
In 1920s the biologist Alexander Gurvich discovered mitogenetic effect: division of cells could be accelerated by optical contact with some biological or chemical system. In his original experiment two onion bulbs were placed distantly and chemically separated, so that only optical contact was possible. He reported increased division intensity in comparison to isolated onion bulbs. There were attempts to explain that by weak ultra-violet signals, but overall reaction to his experiments was controversial. The modern experiments aim to study influence of large number of factors to cell division intensity, so it is close to impossible to process all the data manually. We present an effective and fast algorithm for estimation of division intensity from a set of microscopic images. We test our method on images of budding yeast culture..png)
A cell typically looks like an elliptic spot with a halo around it. The dividing cell could be recognized by a little bud near it..png)
Main steps of the algorithm (left):
- Input image
- Edge detection with the Laplacian of Gaussian filter
- Edges closure, region filling, and median filtration
- Connected components extraction
- Circular approximation (typical radii ranges for cells and buds are specified)
- Cells to buds attribution
Membrane recognition in tissue images
Influence of mechanical factors on cellular tissue organization and organ development is an important problem in biology. There are types of cells that live in a special substrate. The cells interact with the substrate, which influences their further development. For example, transplanted cells may develop into nerves, muscles or connectivity tissues on soft, medium rough, or rough substrate, respectively. To study the influence of mechanical factors on development of the cells and organs, it is essential to have an effective and fast algorithm to parse microscopic images of cellular tissues.
The main problem is to find pixels of membranes, which represent parts of the inter-cellular space. In an image, they are reflected by bands of high brightness between cells.
.png)
We consider the inter-cellular pattern as a graph layout. Thus, nodes are the points where several membranes intersect, and edges (we also refer to them as membranes ) are bands between the cells. Nodes could be recognized as bright spots relatively well, while thresholding works bad for membranes due to volatile lighting conditions. To improve performance we first set low threshold to over-generate nodes, and connect all the neighbouring pairs of candidate nodes with candidate membranes using dynamic programming path finder (the brightest path is selected). Then, a graphical model is constructed to choose optimal consistent configuration of nodes and membranes.
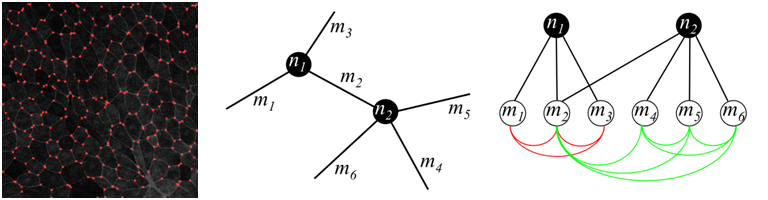
Detected nodes by thresholding on brightness (left); graph over candidate nodes and membranes (center); dependencies of variables in graphical model, red and green lines show two higher-order factors (right)
Each candidate node corresponds to a binary variable ni, each membrane corresponds to a binary variable mj. They are on if and only if a node/membrane is included into final decision. Obviously, each membrane should have nodes in both ends, which places hard constraints on the answer. There are also soft constraints that allow only reasonable number of membranes to meet at a single node. This constraint is expressed in terms of higher-order factors (see figure above).
Papers
- K. Nekrasov, D. Laptev, D. Vetrov. Automatic Detection of Cell Division Intensity in Budding Yeast. Proceedings of International Conference on Pattern Recognition and Image Analysis (PRIA) , vol. 2, pp. 335-339, 2010. pdf
- K. Nekrasov, R. Shapovalov, D. Vetrov. Cellular tissue detection in microscopic images. Technical report, 2012. pdf
Relevant biological work
- Ilya Volodyaev, Ruslan N. Ivanovsky, Elena N. Krasilnikova, Lev V. Beloussov. A partial verification of the A. Gurvich mitogenetic effect. 2009. html
- M. Kvarnstrom, K. Logg, A. Diez, K. Bodvard, M. Kall, Image analysis algorithms for cell contour recognition in budding yeast. Optics express. – 2008. – Vol. 16, No. 17
Have you spotted a typo?
Highlight it, click Ctrl+Enter and send us a message. Thank you for your help!
To be used only for spelling or punctuation mistakes.